Experiment (Count)
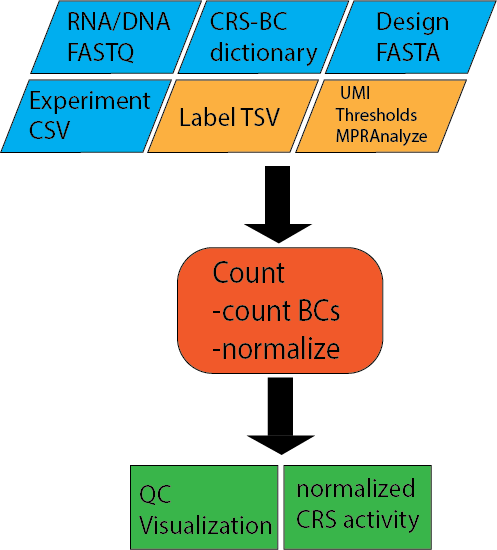
Input files
Experiment File
Comma separated file (CSV) that assigns all fastq files present in a directory to a condidtion and replicate. Each line represents an experiment, which will all be processed in parallel
Condition,Replicate,DNA_BC_F,DNA_UMI,DNA_BC_R,RNA_BC_F,RNA_UMI,RNA_BC_R
Condidtion1,1,C1R1_DNA_barcode_F.fastq.gz,C1R1_DNA_barcode_UMI.fastq.gz,C1R1_DNA_barcode_R.fastq.gz,C1R1_RNA_barcode_F.fastq.gz,C1R1_RNA_barcode_UMI.fastq.gz,C1R1_RNA_barcode_R.fastq.gz
Condidtion1,2,C1R2_DNA_barcode_F.fastq.gz,C1R2_DNA_barcode_UMI.fastq.gz,C1R2_DNA_barcode_R.fastq.gz,C1R2_RNA_barcode_F.fastq.gz,C1R2_RNA_barcode_UMI.fastq.gz,C1R2_RNA_barcode_R.fastq.gz
Condidtion1,3,C1R3_DNA_barcode_F.fastq.gz,C1R3_DNA_barcode_UMI.fastq.gz,C1R3_DNA_barcode_R.fastq.gz,C1R3_RNA_barcode_F.fastq.gz,C1R3_RNA_barcode_UMI.fastq.gz,C1R3_RNA_barcode_R.fastq.gz
Condidtion2,1,C2R1_DNA_barcode_F.fastq.gz,C2R1_DNA_barcode_UMI.fastq.gz,C2R1_DNA_barcode_R.fastq.gz,C2R1_RNA_barcode_F.fastq.gz,C2R1_RNA_barcode_UMI.fastq.gz,C2R1_RNA_barcode_R.fastq.gz
Condidtion2,2,C2R2_DNA_barcode_F.fastq.gz,C2R2_DNA_barcode_UMI.fastq.gz,C2R2_DNA_barcode_R.fastq.gz,C2R2_RNA_barcode_F.fastq.gz,C2R2_RNA_barcode_UMI.fastq.gz,C2R2_RNA_barcode_R.fastq.gz
Condidtion2,3,C2R3_DNA_barcode_F.fastq.gz,C2R3_DNA_barcode_UMI.fastq.gz,C2R3_DNA_barcode_R.fastq.gz,C2R3_RNA_barcode_F.fastq.gz,C2R3_RNA_barcode_UMI.fastq.gz,C2R3_RNA_barcode_R.fastq.gz
Design File
Fasta file of of CRS sequences with unique headers describing each tested sequence
Example file:
>CRS1
GACGGGAACGTTTGAGCGAGATCGAGGATAGGAGGAGCGGA
>CRS2
GGGCTCTCTTATATTAAGGGGGTGTGTGAACGCTCGCGATT
>CRS3
GGCGCGCTTTTTCGAAGAAACCCGCCGGAGAATATAAGGGA
>CRS4
TTAGACCGCCCTTTACCCCGAGAAAACTCAGCTACACACTC
Assignment File or configuration
Tab separated gzipped file with barcode mapped to sequence. Can be generated using the Assignment workflow. Config file must be configured similar to this:
example_assignment:
type: file
value: /path/to/your/file.tsv.gz
Example assignment file:
ATGCGT CRS1
GTCGA CRS2
CCGTT CRS3
CCCCT CRS4
Another option would be referring to an assignment defined in a config file.
example_assignment:
type: config
value: example_config
Label File (Optional)
Tab separated file (TSV) of desired labels for each tested sequence
Example file:
CRS1 Positive_Control
CRS2 Negative_Control
CRS3 Test
CRS4 Positive_Control
Note
If you provide a label file, the first column of the label file must exactly match the FASTA file or the files will not merge properly in the pipeline.
snakemake
Options
With --help or -h you can see the help message.
- Mandatory arguments:
- --cores:
Use at most N CPU cores/jobs in parallel. If N is omitted or ‘all’, the limit is set to the number of available CPU cores. In case of cluster/cloud execution, this argument sets the number of total cores used over all jobs (made available to rules via workflow.cores).(default: None)
- --configfile:
Specify or overwrite the config file of the workflow (see the docs). Values specified in JSON or YAML format are available in the global config dictionary inside the workflow. Multiple files overwrite each other in the given order. Thereby missing keys in previous config files are extended by following configfiles. Note that this order also includes a config file defined in the workflow definition itself (which will come first). (default: None)
- --use-conda:
Required to run MPRAsnakeflow. If defined in the rule, run job in a conda environment. If this flag is not set, the conda directive is ignored. (default: False)
- Recommended arguments:
- --snakefile:
You should not need to specify this. By default, Snakemake will search for ‘Snakefile’, ‘snakefile’, ‘workflow/Snakefile’,’workflow/snakefile’ beneath the current working directory, in this order. Only if you definitely want a different layout, you need to use this parameter. This is very usefull when you want to have the results in a different folder than MPRAsnakeflow is in. (default: None)
- Usefull arguments:
- -n:
Do not execute anything, and display what would be done. If you have a very large workflow, use –dry-run –quiet to just print a summary of the DAG of jobs. (default: False)
- --touch, -t:
Touch output files (mark them up to date without really changing them) instead of running their commands. This is used to pretend that the rules were executed, in order to fool future invocations of snakemake. Fails if a file does not yet exist. Note that this will only touch files that would otherwise be recreated by Snakemake (e.g. because their input files are newer). For enforcing a touch, combine this with –force, –forceall, or –forcerun. Note however that you loose the provenance information when the files have been created in realitiy. Hence, this should be used only as a last resort. (default: False)
Rules
Rules run by snakemake in the assignment utility. Some rules will be run only if certain options used and are marked below.
- create_BAM or create_BAM_noUMI (if no UMI sequence)
creates a bamfile of barcode and UMI sequences
- raw_counts
creates a table of counts for each barcode (where UMIs, if present, are deduplicated)
- filter_counts
Remove barcodes that are not the appropriate length
- final_counts
Record overrepresended UMIs and final count table
- dna_rna_merge_counts or dna_rna_mpranalyze_merge
Merge RNA/DNA count matrices per barcode
- final_merge (MPRAnalyze option only)
Merge all DNA/RNA counts into one file
- final_label (MPRAnalyze option only)
Label the barcodes
- generate_mpranalyze_inputs (MPRAnalyze option only)
Generate inputs for MPRAnalyze, counts tables and annotation tables for rna/dna
- dna_rna_merge
Merge each DNA and RNA file label with sequence and insert and normalize
- calc_correlations
Calculate correlations between Replicates
- make_master_tables
Create tables of each CRS normalized across replicates
Output
The output can be found in the folder defined by the option results/experiments/. It is structured in folders of the condition as
Files
File tree
experimet_name
|-Condition
|-allreps.tsv
|-average_allreps.tsv
|-HepG2_1_2_correlation.txt
|-HepG2_1_2_DNA_pairwise.png
|-HepG2_1_2_Ratio_pairwise.png
|-HepG2_1_2_RNA_pairwise.png
|-HepG2_all_barcodesPerInsert_box.png
|-HepG2_barcodesPerInsert.png
|-Reps
|-HepG2_1_counts.tsv
|-HepG2_1_counts.tsv.gz
|-HepG2_1_DNA_counts_full.tsv
|-HepG2_1_DNA_counts_full_samplingN.tsv
|-HepG2_1_DNA_raw_counts.tsv.gz
|-HepG2_1_RNA_filtered_counts.tsv.gz
|-HepG2_1_DNA_filtered_counts.tsv.gz
|-HepG2_1_RNA_counts.tsv
|-HepG2_1_RNA_raw_counts.tsv.gz
Todo
This is not the correct file tree for the experiment workflow
Files for each Condition
- allreps.tsv
TSV of normalized DNA and RNA count, ratio, log2ratio, and number of observed barcodes for each condition, replicate, of every CRS
- average_allreps.tsv
mean ratio, log2 ratio, and observed barcodes per condidition normalized for all replicates
- HepG2_1_2_correlation.txt
correlation values for a condition and 2 replicates (ie: HepG2 replicate 1 vs replicate 2)
- HepG2_1_2_DNA_pairwise.png
Correlation plot of DNA counts condition vs two reps (ie: HepG2 replicate 1 vs replicate 2)
- HepG2_1_2_Ratio_pairwise.png
Correlation plot of normalized log2(RNA/DNA) condition vs two reps (ie: HepG2 replicate 1 vs replicate 2)
- HepG2_1_2_RNA_pairwise.png
Correlation plot of RNA counts condition vs two reps (ie: HepG2 replicate 1 vs replicate 2)
- HepG2_all_barcodesPerInsert_box.png
Box plot of each CRS accross replicates for all barcodes in each condidtion. Colored by the label file.
- HepG2_barcodesPerInsert.png
Histogram of number of barcodes detected per CRS
- HepG2_group_barcodesPerInsert_box.png
Boxplot of CRS normalized per insert, grouped by labels
Todo
These are not the correct files for each condition in the experiment workflow
Files for each replicate in each condition
- HepG2_1_counts.tsv
mean ratio, log2 ratio, and observed barcodes per condidition for each replicate
- HepG2_1_counts.tsv.gz
table of barcodes with DNA counts and RNA counts
- HepG2_1_DNA_counts_full.tsv
table of barcodes with DNA counts
- HepG2_1_DNA_counts_full_samplingN.tsv
table of barcodes with DNA counts with adjusted sampling.
- HepG2_1_DNA_raw_counts.tsv.gz
table of barcodes, UMI, and DNA counts raw
- HepG2_1_DNA_filtered_counts.tsv.gz
table of barcodes, UMI, and DNA counts raw, filtered for barcodes of correct length
- HepG2_1_RNA_counts.tsv
table of barcodes with RNA counts
- HepG2_1_RNA_raw_counts.tsv.gz
table of barcodes, UMI, and RNA counts raw
- HepG2_1_RNA_filtered_counts.tsv.gz
table of barcodes, UMI, and DNA counts raw, filtered for barcodes of correct length
Todo
These are not the correct files for the experiment workflow